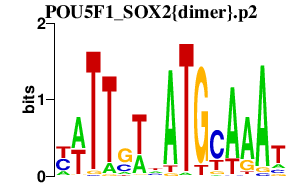
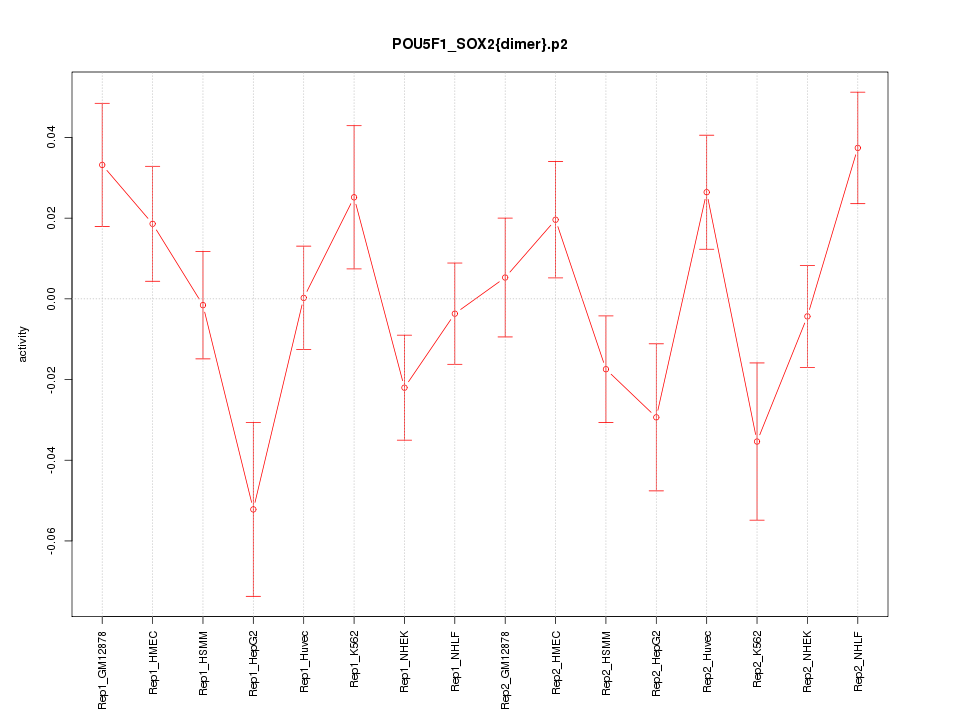
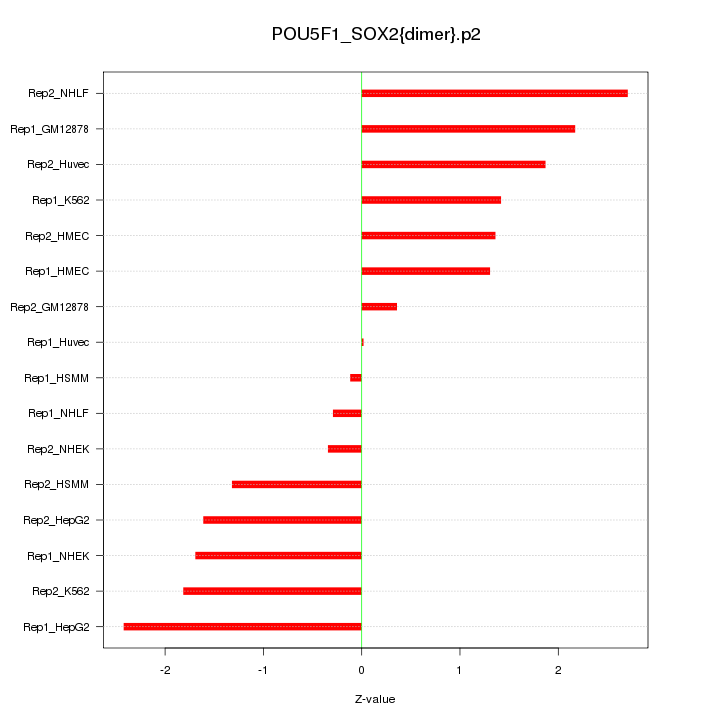
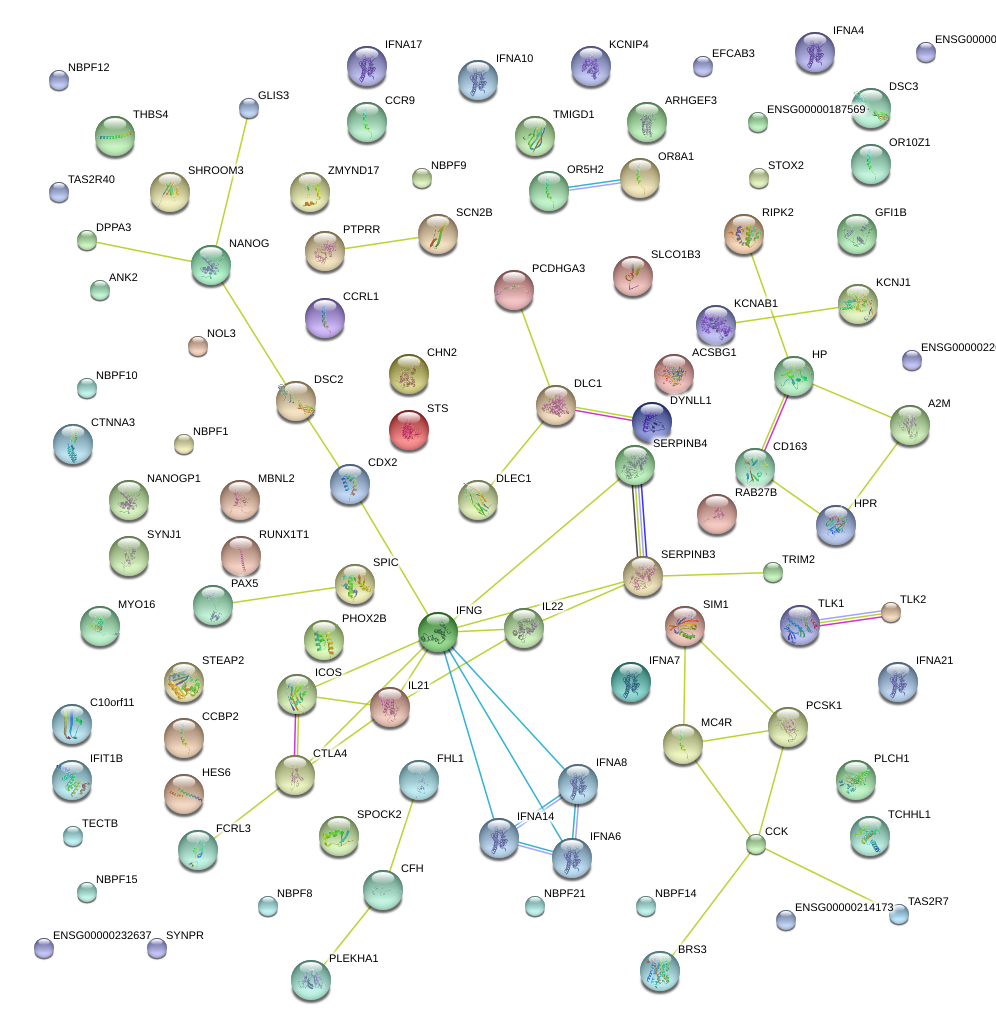
|
chr5_-_95794416
|
2.287
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr10_+_114033403
|
1.750
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr4_+_77575276
|
1.741
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr18_-_56190980
|
1.706
|
NM_005912
|
MC4R
|
melanocortin 4 receptor
|
|
chr7_+_89681069
|
1.635
|
NM_001040666
|
STEAP2
|
six transmembrane epithelial antigen of the prostate 2
|
|
chr10_-_73518772
|
1.607
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr10_+_77212524
|
1.564
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr12_-_10846435
|
1.495
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr12_-_66839589
|
1.488
|
NM_000619
|
IFNG
|
interferon, gamma
|
|
chr1_-_150328153
|
1.479
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr7_+_29486010
|
1.471
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr3_-_156904690
|
1.419
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr4_+_185063421
|
1.293
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr3_-_42280442
|
1.289
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr9_-_4142182
|
1.286
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_+_204509698
|
1.191
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr13_-_27441290
|
1.177
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr1_-_155936904
|
1.169
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr17_-_25685175
|
1.129
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr3_+_157491469
|
1.124
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr17_+_57811645
|
1.092
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr4_+_114433551
|
1.084
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr12_+_7755316
|
1.056
|
NM_199286
|
DPPA3
STELLAR
|
developmental pluripotency associated 3
developmental pluripotency associated 3 pseudogene
|
|
chr1_+_156842852
|
1.050
|
NM_001004478
|
OR10Z1
|
olfactory receptor, family 10, subfamily Z, member 1
|
|
chr9_-_21177529
|
1.047
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr2_-_238813365
|
1.030
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr13_-_27441143
|
1.030
|
|
CDX2
|
caudal type homeobox 2
|
|
chr9_+_21399132
|
1.027
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr10_+_91127792
|
1.018
|
NM_001010987
|
IFIT1B
|
interferon-induced protein with tetratricopeptide repeats 1B
|
|
chr6_-_101018271
|
1.006
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr5_+_79366746
|
0.995
|
NM_003248
|
THBS4
|
thrombospondin 4
|
|
chr9_-_21340885
|
0.972
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr3_+_45902999
|
0.958
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr11_+_123945102
|
0.957
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr7_+_142629251
|
0.955
|
NM_176882
|
TAS2R40
|
taste receptor, type 2, member 40
|
|
chr11_-_117552545
|
0.955
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr12_+_7833261
|
0.921
|
NM_024865
|
NANOG
|
Nanog homeobox
|
|
chr13_-_27441423
|
0.911
|
|
CDX2
|
caudal type homeobox 2
|
|
chr5_+_140751666
|
0.910
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr2_-_238813284
|
0.900
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr15_-_76313896
|
0.899
|
NM_015162
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr9_+_134843918
|
0.892
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr9_-_21229975
|
0.875
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr9_-_21197141
|
0.861
|
NM_002171
|
IFNA10
|
interferon, alpha 10
|
|
chr10_-_74863303
|
0.823
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr13_+_108079570
|
0.821
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr12_-_7547552
|
0.820
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr3_+_99484421
|
0.819
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chr18_-_59462469
|
0.814
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chrX_+_135106578
|
0.809
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_+_7833292
|
0.774
|
|
NANOG
|
Nanog homeobox
|
|
chr3_+_63403792
|
0.764
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr12_+_20854904
|
0.759
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr12_+_103221590
|
0.755
|
NM_001008394
|
EID3
|
EP300 interacting inhibitor of differentiation 3
|
|
chr18_+_50646616
|
0.753
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr1_-_146492471
|
0.751
|
NM_015383
|
NBPF14
NBPF1
|
neuroblastoma breakpoint family, member 14
neuroblastoma breakpoint family, member 1
|
|
chr4_-_21155376
|
0.750
|
NM_001035004
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr12_-_9123126
|
0.747
|
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_+_135397790
|
0.744
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr7_+_37689903
|
0.730
|
|
|
|
|
chr10_-_69125932
|
0.721
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr18_-_26876622
|
0.719
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr13_+_96592051
|
0.717
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr9_-_21192201
|
0.713
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr10_+_124141808
|
0.712
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chrX_+_7147471
|
0.700
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr2_-_171631728
|
0.693
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr4_-_123761615
|
0.691
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr8_-_93099040
|
0.684
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_140719773
|
0.676
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr4_+_154397975
|
0.672
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr12_-_69600514
|
0.663
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr4_-_41445479
|
0.661
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr3_-_56784634
|
0.658
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr11_-_128242342
|
0.648
|
NM_153764
NM_153765
NM_153766
NM_153767
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr16_+_70646008
|
0.639
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr1_+_190552744
|
0.637
|
NM_001039152
|
RGS21
|
regulator of G-protein signaling 21
|
|
chr6_-_76838944
|
0.635
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr6_+_50894397
|
0.635
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr9_+_6205804
|
0.633
|
|
IL33
|
interleukin 33
|
|
chr13_+_22653059
|
0.623
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr3_+_109504021
|
0.621
|
NM_007072
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr8_-_26780645
|
0.617
|
|
|
|
|
chr12_+_102859354
|
0.614
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr1_+_116181927
|
0.612
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr5_-_135318420
|
0.601
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr2_-_215998181
|
0.601
|
|
FN1
|
fibronectin 1
|
|
chrX_-_55037235
|
0.600
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr7_-_107230867
|
0.595
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr16_+_74868676
|
0.594
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr1_-_93172119
|
0.591
|
|
|
|
|
chr4_+_14950657
|
0.586
|
NM_001135170
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr3_+_99555387
|
0.584
|
NM_001005517
|
OR5K4
|
olfactory receptor, family 5, subfamily K, member 4
|
|
chrX_-_15312390
|
0.581
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr10_+_123744131
|
0.575
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chrX_+_135210787
|
0.572
|
NM_153834
|
GPR112
|
G protein-coupled receptor 112
|
|
chr16_+_52081836
|
0.571
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr3_-_38810504
|
0.570
|
NM_006514
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit
|
|
chr3_+_12367993
|
0.567
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr7_+_129057152
|
0.565
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr2_-_215949566
|
0.562
|
|
FN1
|
fibronectin 1
|
|
chr3_+_149930656
|
0.558
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr11_-_117552355
|
0.557
|
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr12_-_88573949
|
0.554
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr1_-_108588186
|
0.553
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chrX_+_70255128
|
0.551
|
NM_005120
|
MED12
|
mediator complex subunit 12
|
|
chr9_-_21156609
|
0.544
|
NM_002175
|
IFNA21
|
interferon, alpha 21
|
|
chr3_-_42892195
|
0.541
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr17_+_29606408
|
0.539
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr11_+_113280727
|
0.537
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr14_-_20098929
|
0.536
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr14_+_61532293
|
0.529
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr2_-_37397669
|
0.528
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr8_-_26780665
|
0.526
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr1_-_3556420
|
0.516
|
NM_017818
|
WDR8
|
WD repeat domain 8
|
|
chr10_-_52315436
|
0.514
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr5_+_140762551
|
0.510
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr2_-_20390601
|
0.508
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr14_-_106149623
|
0.503
|
|
|
|
|
chr1_-_229071957
|
0.498
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr1_+_86785346
|
0.491
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr11_-_57171554
|
0.491
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_+_99370233
|
0.487
|
NM_001005515
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15
|
|
chr2_-_200038055
|
0.486
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr12_+_54231249
|
0.485
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chr4_+_74493918
|
0.485
|
|
ALB
|
albumin
|
|
chr1_-_41398126
|
0.484
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr6_-_32605898
|
0.482
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr1_-_175400441
|
0.474
|
|
ASTN1
|
astrotactin 1
|
|
chr15_+_19883745
|
0.474
|
NM_001005241
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4
|
|
chr4_-_41445693
|
0.473
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr9_+_91409746
|
0.470
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr4_-_165338312
|
0.469
|
NM_012403
|
ANP32C
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member C
|
|
chr6_-_89984214
|
0.468
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr12_+_54172413
|
0.466
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr12_-_100128115
|
0.462
|
NM_145913
|
SLC5A8
|
solute carrier family 5 (iodide transporter), member 8
|
|
chrX_+_141941369
|
0.455
|
NM_001009613
|
SPANXN4
|
SPANX family, member N4
|
|
chr8_+_7340777
|
0.451
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr15_+_63701322
|
0.447
|
NM_004727
|
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1
|
|
chr4_-_74307637
|
0.446
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr3_+_99350859
|
0.446
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chrX_-_138875342
|
0.443
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr4_+_120276386
|
0.439
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr2_-_45648608
|
0.437
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr8_+_142501188
|
0.437
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr8_-_7297296
|
0.434
|
NM_058206
NM_058207
|
SPAG11B
|
sperm associated antigen 11B
|
|
chr12_-_21379098
|
0.434
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr1_-_175400573
|
0.430
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr10_+_124660206
|
0.430
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr7_-_122126422
|
0.430
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr3_-_174341653
|
0.424
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr10_-_5436781
|
0.423
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr11_-_102100833
|
0.423
|
NM_002424
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase)
|
|
chr2_+_89715037
|
0.420
|
|
|
|
|
chr3_-_110155237
|
0.416
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr12_+_54106304
|
0.413
|
NM_001005183
|
OR6C76
|
olfactory receptor, family 6, subfamily C, member 76
|
|
chr9_-_21132143
|
0.413
|
NM_002177
|
IFNW1
|
interferon, omega 1
|
|
chr2_-_165768798
|
0.413
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr3_+_2908920
|
0.412
|
NM_175613
|
CNTN4
|
contactin 4
|
|
chr12_+_47162552
|
0.410
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr10_-_7699192
|
0.409
|
|
ITIH5
|
inter-alpha (globulin) inhibitor H5
|
|
chr11_-_57927457
|
0.406
|
NM_001005469
|
OR5B3
|
olfactory receptor, family 5, subfamily B, member 3
|
|
chr11_-_35398077
|
0.405
|
NM_001195728
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr18_-_59480082
|
0.400
|
NM_006919
|
SERPINB3
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr4_-_149577404
|
0.400
|
|
|
|
|
chr8_+_120148604
|
0.399
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr7_-_16811131
|
0.392
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr11_-_58368555
|
0.391
|
NM_145016
|
GLYATL2
|
glycine-N-acyltransferase-like 2
|
|
chr6_-_22410993
|
0.387
|
NM_001163558
|
PRL
|
prolactin
|
|
chr1_+_93072947
|
0.387
|
|
RPL5
|
ribosomal protein L5
|
|
chr15_+_36014748
|
0.386
|
NM_152453
|
TMCO5A
|
transmembrane and coiled-coil domains 5A
|
|
chr11_+_59804634
|
0.383
|
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chr4_-_151993422
|
0.381
|
|
|
|
|
chr1_-_161380562
|
0.381
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr13_+_112746908
|
0.380
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr18_+_41448763
|
0.379
|
NM_007163
|
SLC14A2
|
solute carrier family 14 (urea transporter), member 2
|
|
chr1_-_157772420
|
0.377
|
NM_001004469
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5
|
|
chr5_-_58331515
|
0.377
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_55489973
|
0.376
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr10_+_90552466
|
0.376
|
NM_001128215
|
LIPM
|
lipase, family member M
|
|
chr14_-_57030227
|
0.374
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr3_+_191588354
|
0.374
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr6_-_119512035
|
0.373
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr7_+_45899621
|
0.372
|
|
|
|
|
chrX_+_54483565
|
0.371
|
NM_058163
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr11_-_19038803
|
0.368
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr22_+_34025267
|
0.368
|
NM_001135729
|
TOM1
|
target of myb1 (chicken)
|
|
chr13_-_43101608
|
0.365
|
NM_001127615
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr11_-_59390518
|
0.364
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr10_+_95838801
|
0.361
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr4_+_88939725
|
0.359
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr12_+_129212955
|
0.357
|
NM_007197
|
FZD10
|
frizzled homolog 10 (Drosophila)
|
|
chr18_+_61568825
|
0.355
|
|
CDH7
|
cadherin 7, type 2
|
|
chrX_+_10084984
|
0.352
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr9_-_111230926
|
0.352
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr1_-_116185215
|
0.351
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr7_-_122422989
|
0.350
|
NM_016945
|
TAS2R16
|
taste receptor, type 2, member 16
|
|
chr15_-_39151468
|
0.347
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr7_-_100092003
|
0.343
|
NM_016188
|
ACTL6B
|
actin-like 6B
|
|
chr4_-_154929657
|
0.339
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|